Bioinformatics
Bioinformatics services
Since 2007, the Rnomics Platform provides bioinformatics services to Sherbrooke University community and to canadian and external academics. We can assist you in the analysis of your sequencing data and the production of publication quality figures. We also offer a robust and exhaustive PCR primer design service. Hourly rate and accessibility are eased for members of the Sherbrooke University community.
Sequencing data analysis
We offer a extensive panel of pipelines and workflows, addressing several fields of applications. We are always available to discuss about what would be the best way to adapt a workflow to your project, and help you understand each step toward the final results. Here are some applications:
| Variant calling and annotation |
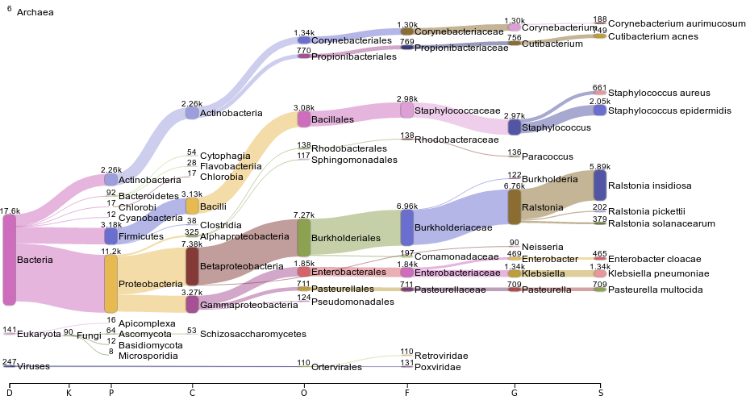
| Metagenomics characterization by 16S or whole genome |
| Bacterial genome and virome reconstruction |
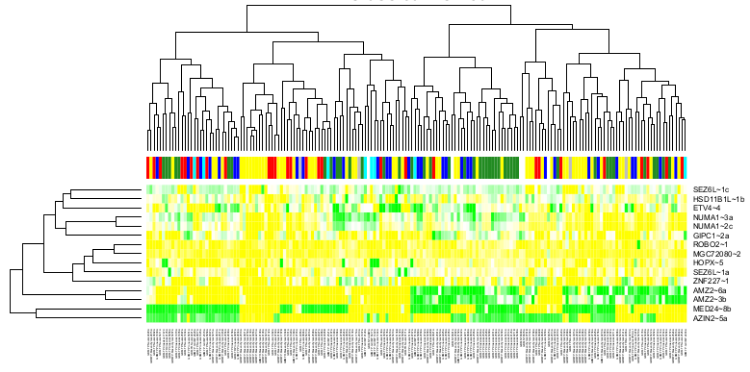
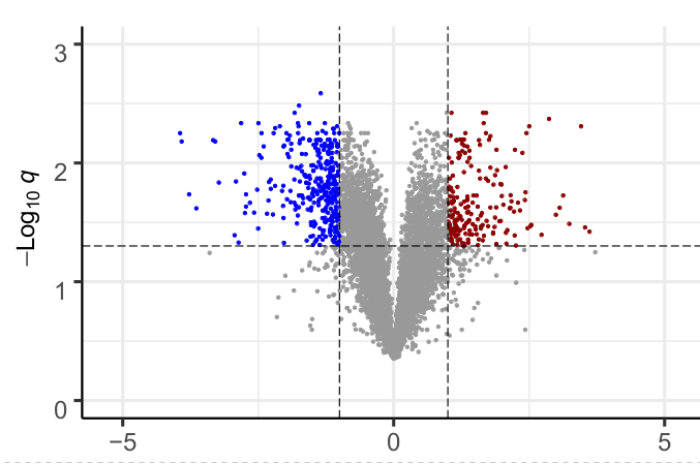
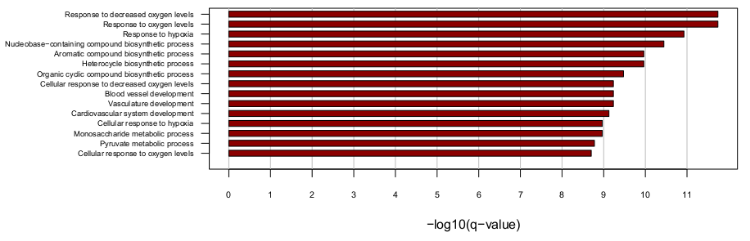
| Gene and isoform expression analysis |
| Alternative splicing analysis |
PCR primer design
We have a robust and scalable PCR primer design service for any target or species. We can target at the gene or isoform level. Available applications:
| Gene or isoform expression by qPCR or ddPCR |
| Alternative splicing event detection and quantification by end-point PCR |
| Bacterial or viral strain detection |
| Genotyping |
Development
We are always open to develop new bioinformatics applications tailored to fit in your research project. We can adapt existing bioinformatics tools, or develop new software to meet your needs. Please contact us for more information.
| Hourly rates CAD$ | |||
|---|---|---|---|
| USherbrooke | Can academic | ||
| 75 | 112.50 | ||
- Rates subject to change.